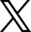For a downloadable PDF, please click here.
About the Gemini® APU
The Associative Processing Unit (APU) is GSI’s patented processing technology and a new breed of processor. It features massive parallel data processing, compute and search, in-place, directly in the memory array. The APU’s design eliminates bandwidth-costly data transfers between the memory and processor. This gives the APU a performance edge in the acceleration of similarity search applications.
The APU as a Driver of Similarity Search
Similarity search is the most general term used for a range of mechanisms which share the principle of searching (typically, very large) spaces of objects where the only available comparator is the similarity between any pair of objects. This is becoming increasingly important in an age of large information repositories where the objects contained do not possess any natural order.
Similarity search involves searching a database of vectors for similarity to a given query. The query itself must be of identical size to the database records (e.g., 512 or 1024 bits).
At GSI, we implement similarity search and top-k using parallel processing. Current processor technologies are not well suited to similarity search and top-k problems due to the high memory bandwidth demands required. For similarity search. the APU’s advantage over the CPU is processing time—the APU can process a similarity search in less than 15 microseconds, with lower power consumption and latency.
Similarity Structure Search in Cheminformatics
Cheminformatics is concerned with the development of data formats and databases containing information about different aspects of various chemical systems, and the design of tools to query those databases. Cheminformatics aims to create a user interface that assembles the different tools needed to query data as a whole. Among the main objectives of cheminformatics, are:
- Reduction of the cost and time needed to develop new drugs
- Increasing the efficacy and properties of natural substances
- Production of new protein molecules and chemical materials
- Genome sequencing
Cheminformatics uses very large databases. To date, several databases have been developed for cataloging molecule data for drug development.
These databases consist of chemical fingerprints, used to identify the features of chemical elements within a matrix, and therefore define its unique portrait in comparison to similar matrices. Molecular fingerprints have been used for a long time now in drug discovery and virtual screening. Their ease of use (requiring little to no configuration) and the speed at which substructure and similarity searches can be performed with them—paired with a virtual screening performance like other more complex methods—is the reason for their popularity.
GSI Joins Forces with the Weizmann Institute of Science and G-INCPM
Project Overview
As a proof of concept for the APU technology with similarity search applications, GSI Technology has teamed up with researchers at the Weizmann Institute of Science, in Rehovot, Israel, and The Nancy & Stephen Grand Israel National Center for Personalized Medicine (G-INCPM). The Weizmann Institute has a world class reputation in multidisciplinary scientific research, while G-INCPM is an advanced research facility that strives to promote the field of personalized medicine.
As part of ongoing research on new molecules with pharmacological properties, and with the goal of obtaining the approval of the FDA and other agencies, the team at the Weizmann Institute and G-INCPM is searching an established database of 38 million molecular fingerprints with the goal of identifying molecules that are structurally similar—or have similar properties—to their query molecule.
The Weizmann Institute of Science & G-INCPM’s Approach to Structure Search
Researchers at the Weizmann Institute and G-INCPM have been using BIOVIA Pipeline Pilot[1] data analysis software, Oracle Database, and a CPU to fetch, process, and analyze molecular structures data. However, they are finding that fetching the data with Oracle Database and performing similarity structure search with a CPU is a relatively slow and inefficient process.
A New Approach with Gemini® APU
To speed up the similarity structure search process, GSI is replacing the Oracle-based BIOVIA Tanimoto search engine with an APU-based Tanimoto search engine. GSI has replaced the BIOVIA DIRECT search component in Pipeline Pilot that calls Oracle Database and supports only MDL Public keys fingerprint type, with a new Python API that calls the APU processor and the GSI Search Library (GSL). In the GSL, GSI has implemented similarity structure search based on the Tanimoto distance measures.
The architecture diagram provides an overview of GSI’s APU similarity structure search solution. The GSI Fast Vectors search API connects Pipeline Pilot to GSL. It can equally be used by other third-party applications, such as KNIME, and Search by Elastic Search Plugin that connect to the GSI Fast Vector Search API.
The GSI Similarity Structure Search Process
- The binary database (Covert to Binary by GSI Tool) is loaded ONCE into the L4[2] buffer. It will only need to be loaded again if changes are made to it. This step represents a new capability in Pipeline Pilot, developed by GSI.
- Pipeline Pilot converts the researcher’s query molecule into a folded fingerprint string type ECFP/FCFP. (Existing functionally.)
- GSI replaces the existing Pipeline Pilot search block with a GSI Search block. This block calls the GSI Search Library (GSL) function that performs the following actions:
- The query is loaded into the L1 memory block.
- The molecule database is divided into manageable chunks of data that can fit into L1[3] memory.
- We use the Tanimoto coefficient to perform similarity search. It is considered an ideal method for fingerprint-based calculations of the similarity of molecular representations.
- GSI’s Python code calls the GNL Tanimoto distance measures, which find values and indices of k nearest entries.
[2] L4 is the APU’s DDR4 DRAM system memory, used to store the vector database of 38 million compounds.
[3] L1 is the vector memory of the APU. It serves as a closely-coupled data storage/cache to the Main Memory Block (MMB)—where data processing takes place.
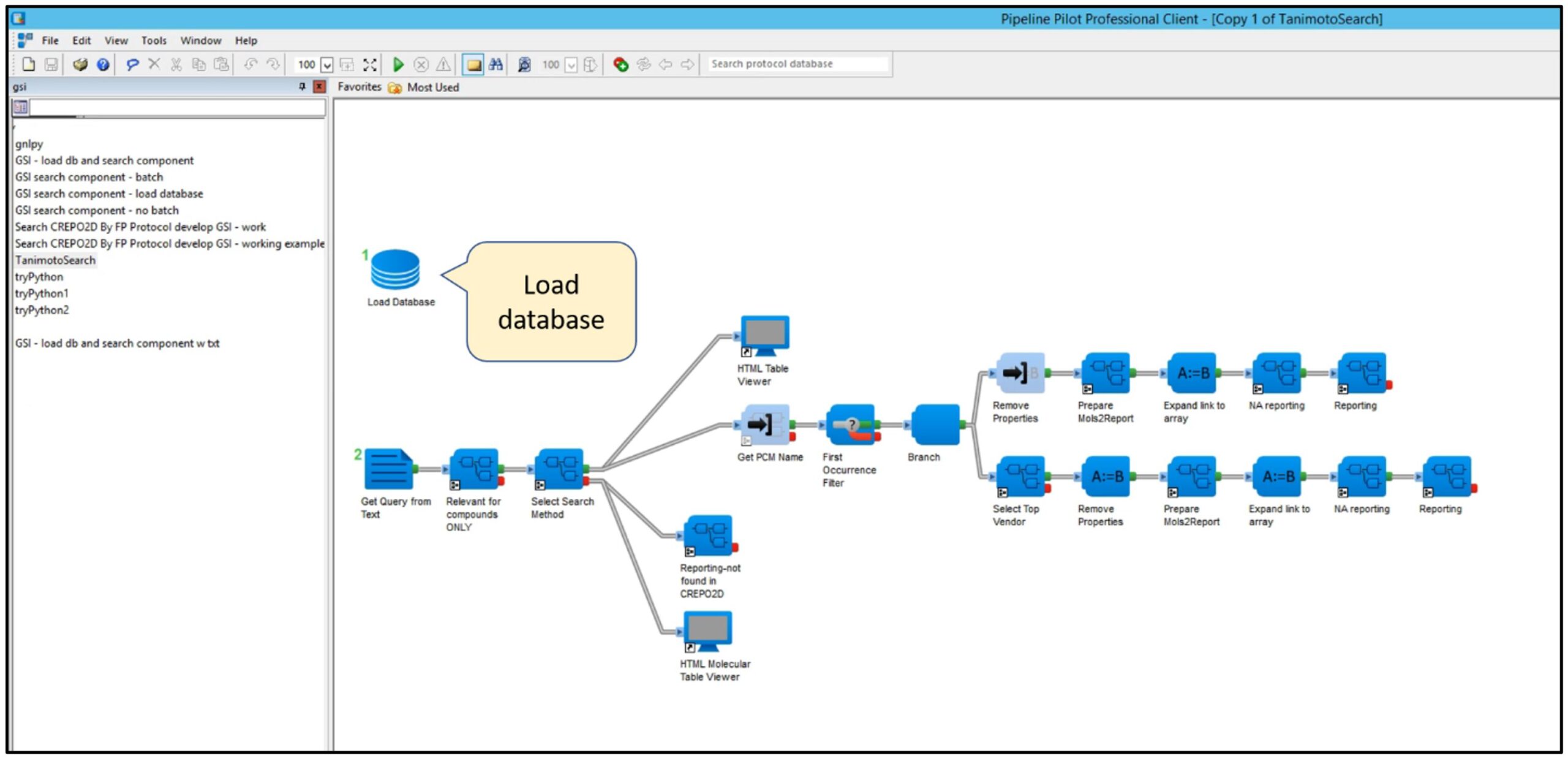
Some Screenshots From Pipeline Pilot
The following screenshots from BIOVIA’s Pipeline Pilot user interface show the similarity search pipeline, including the steps where GSI’s Python API code has been embedded in the pipeline, connecting GSI’s APU and GSL resources.
Step 1: Load database.
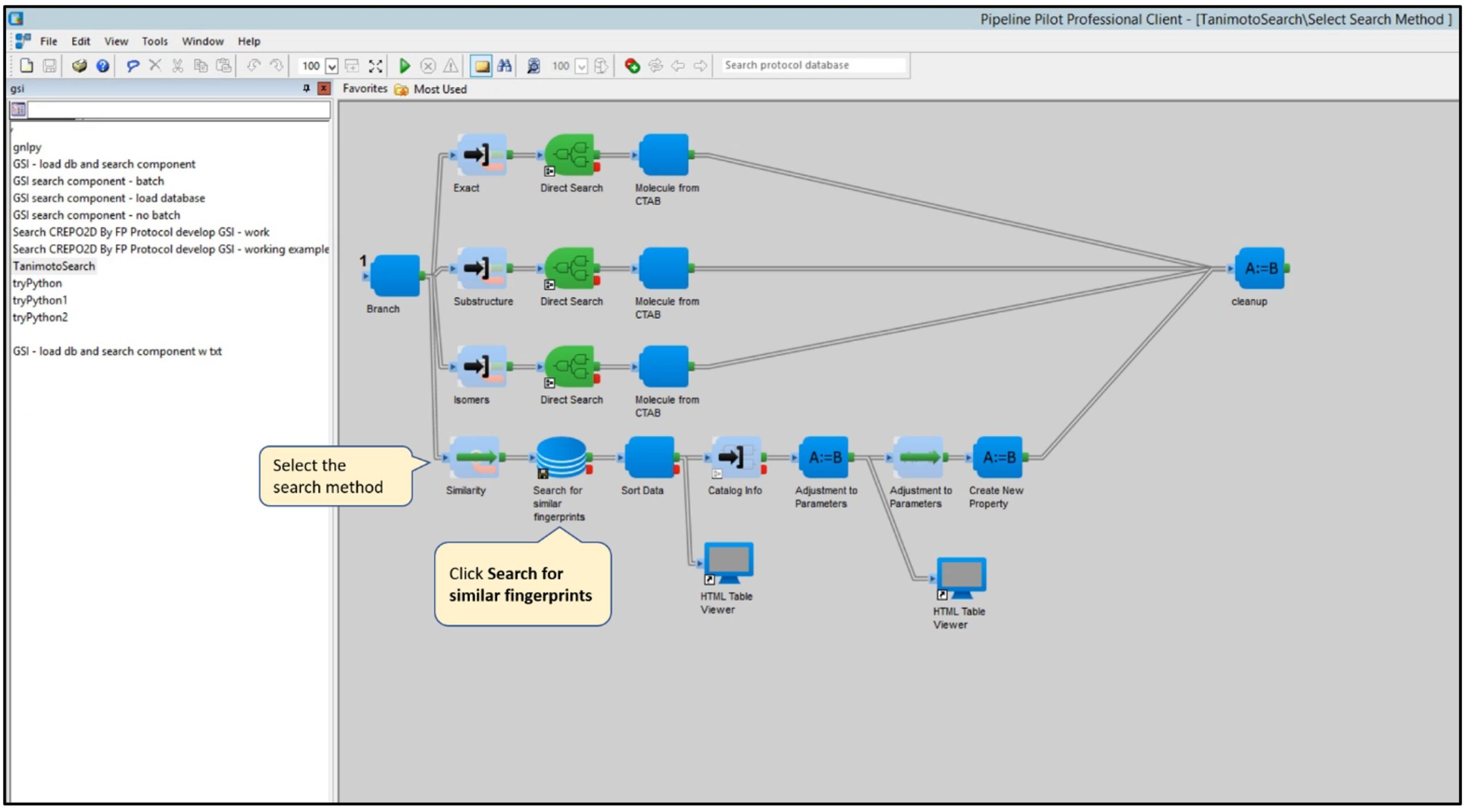
Step 2: Select the search method (e.g., Tanimoto distance) and click Search for similar fingerprints.
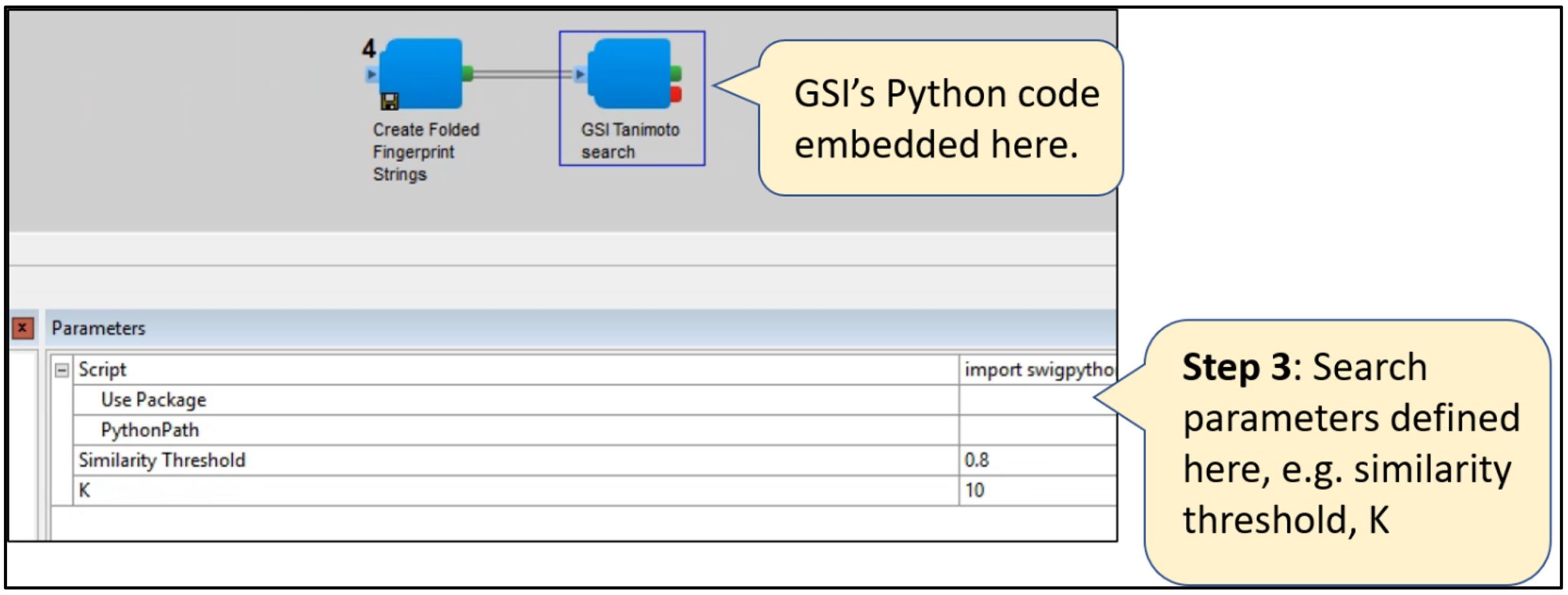
Step 3: Define the search parameters such as K, or similarity Tanimoto threshold.
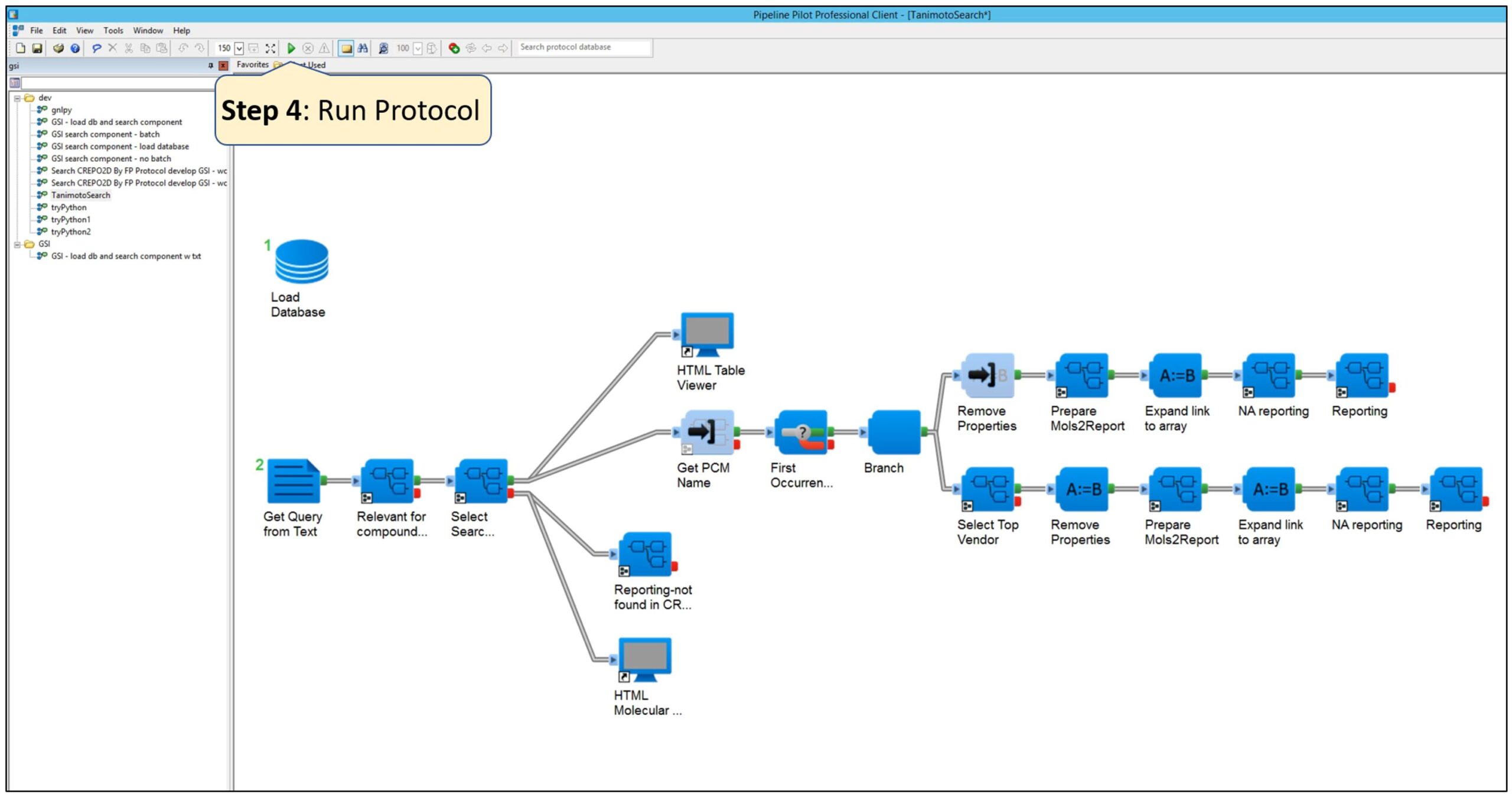
Step 4: Run the protocol.
Performance and Power Consumption
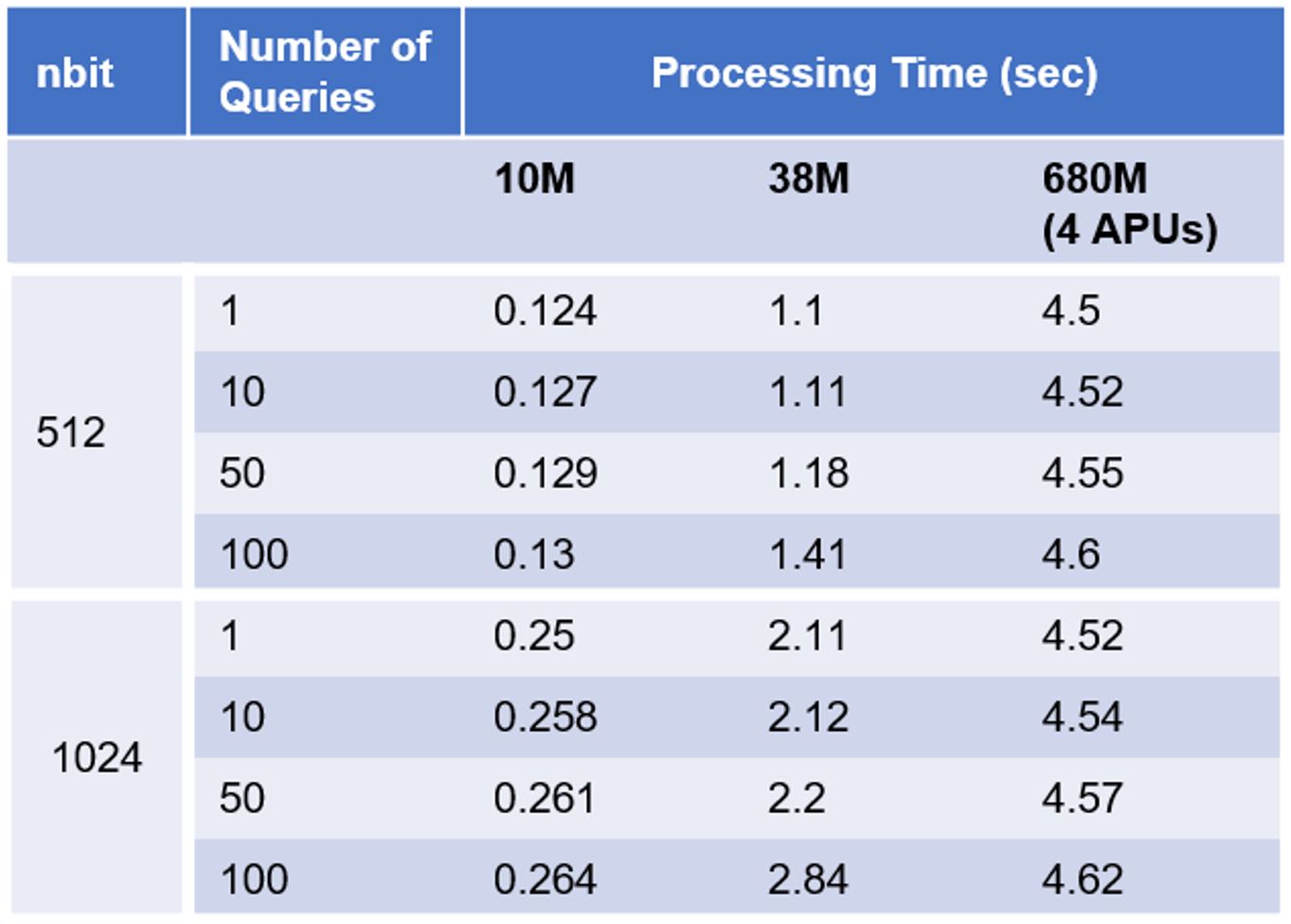
The following table shows Gemini® APU query processing time for KNN similarity search, with 512-bit and 1024-bit vectors, 3 database sizes, and number of queries ranging from 1 to 100.
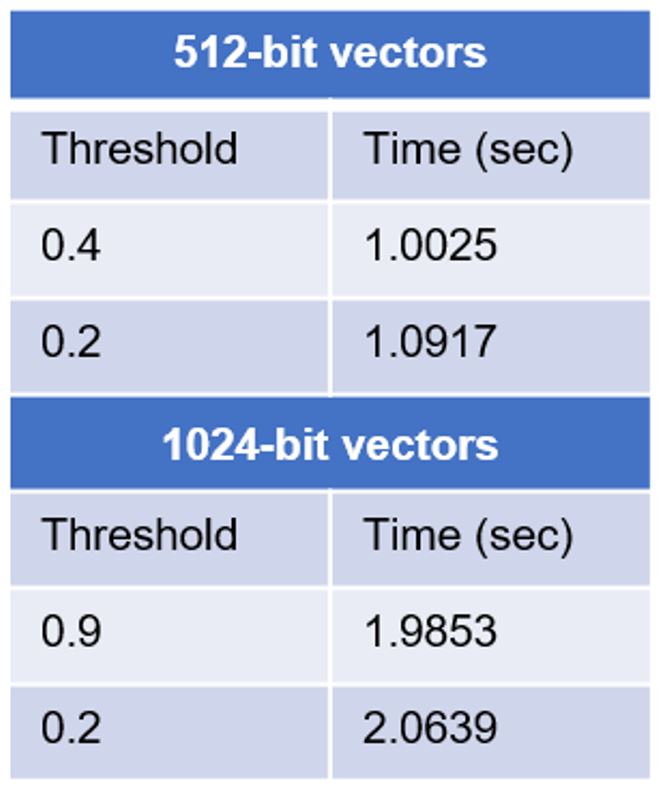
The table below shows selected results for query processing on a 38M database, using distance threshold similarity search.
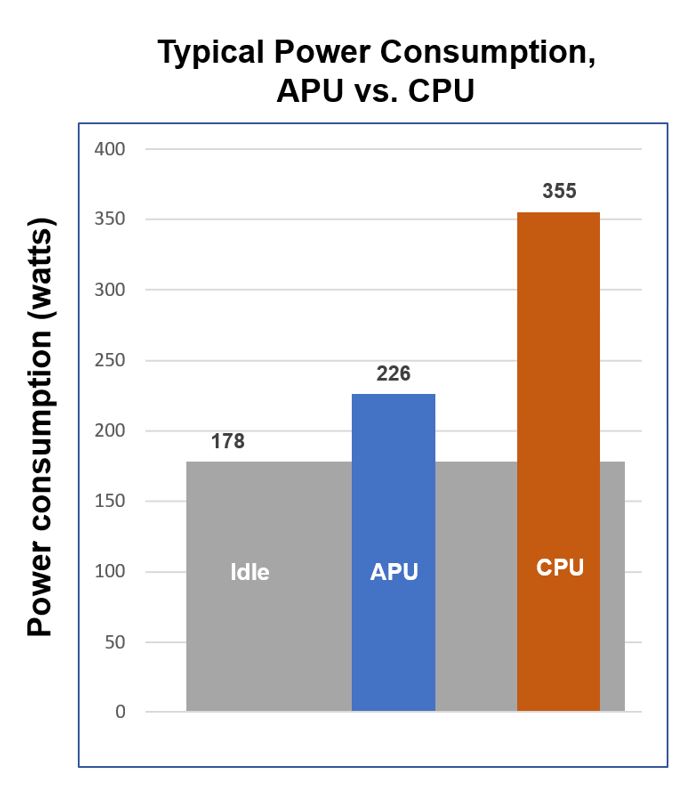
Power Consumption
The APU reduces search power consumption by a factor of 3.5.
Gemini APU Benefits
- Support for similarity structure search on circular fingerprints at thresholds of 0.8 and under.
- Easy integration with BIOVIA – proven and fully tested.
- Gemini® can be integrated into other molecule search solutions that use the Tanimoto coefficient.
- Support for single or multiple queries on Enamine REAL database (680M compounds), all on a single APU.
- Calculation of more descriptive, larger length, folded fingerprints (bit sizes of 256, 512, 1024, and up to 8192).
- Multiple databases can be loaded into the APU’s system memory. Single dataset can be chosen for a specific query.
- Batches of compounds (100s or 1000s) can be submitted simultaneously.
Future Implementations
The APU can be easily integrated with other similarity search applications, using a GSI Fast Vector search (Python or RestAPI) API.